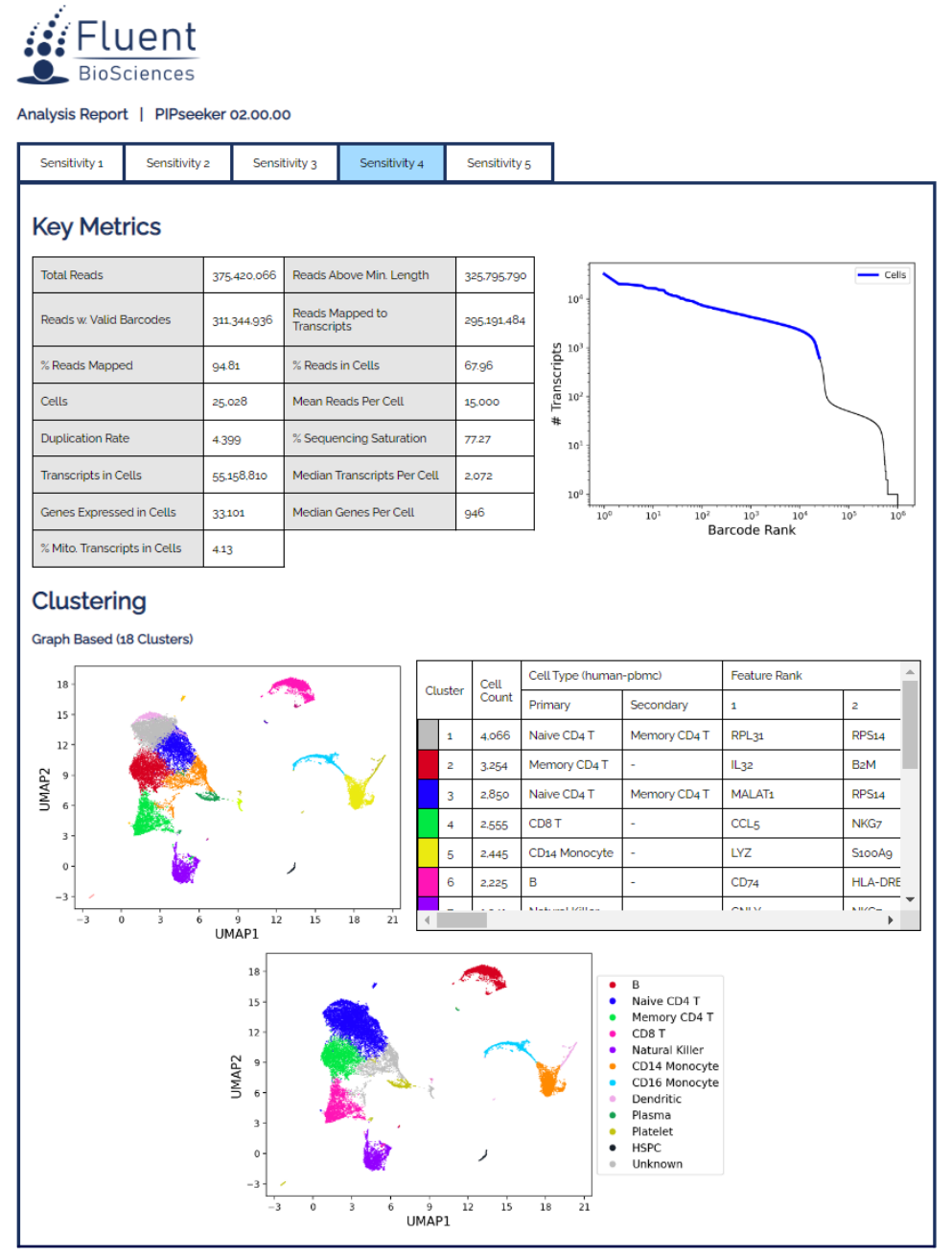
PIPseeker™ Software for Single Cell Data Analysis
- Analyze sequencing data from Illumina 3′ Single Cell RNA Prep Gene Expression libraries
- Obtain summary metrics, diagnostic plots, clustering and differential gene expression tables
- Generate standard feature-barcode count matrices compatible with standard tertiary analysis packages
- Compatible with Linux, macOS, and Windows
Downloads
PIPseeker is available for download as a self-contained package. Common reference packages are also provided in the downloads page, and users can generate their own references as needed.
